TCR Repatoir Analysis
The TCR Repathore is an "immune memory barcode" that reflects an individual's history of antigen sensitization and the individual's ability to respond to various pathogens.
T cells express T cell receptors (TCRs) that recognize antigens from pathogenic microorganisms, cancer, etc. TCR genes are genetically rearranged to produce a vast array of types, and T cells with the same type of TCR gene are called clones. When clonal proliferation occurs in cancer, infectious diseases, autoimmune diseases, etc., in which specific T cell clones recognize antigens and proliferate, a population with a high frequency of a particular type of clone is formed. The collection of TCR genes (clone types and frequency of individual clones) that T cells have as a population is called TCR repatria, and TCR repatria analysis is a method of analyzing this collection using next-generation DNA sequencers and other devices.
As an "immune memory barcode" that reflects an individual's history of antigen sensitization, and as an indicator of an individual's ability to respond to a variety of pathogens, the TCR Repathore has potential applications in the study and diagnosis of cancer, autoimmune diseases, infectious diseases, and vaccine responses.
TCR gene reconstitution in the thymus and clonal selection
Functional and safe clone selection through positive and negative selection
Clonal growth of clones in peripheral tissues in response to pathogens, cancer, self-antigens, etc.
TCR Repertoire consisting of various clones with different frequencies and recognition antigens
High sensitivity, Unbiased
By synthesizing cDNA from mRNA and amplifying the TCR gene with a common primer, IGT's amplification method is more sensitive and unbiased than systems that start with cDNA and amplify the variable region with multiple primers. Moreover, the TCR gene expression level of quiescent T cells is low (a few copies per cell), and repertoireanalysis requires high detection sensitivity. IGT's amplification method is one of the most sensitive among existing technologies.
Duplicate clonal analysis of tumor-infiltrating T cells
CD4/CD8 repertoire analysis in cancer tissue:
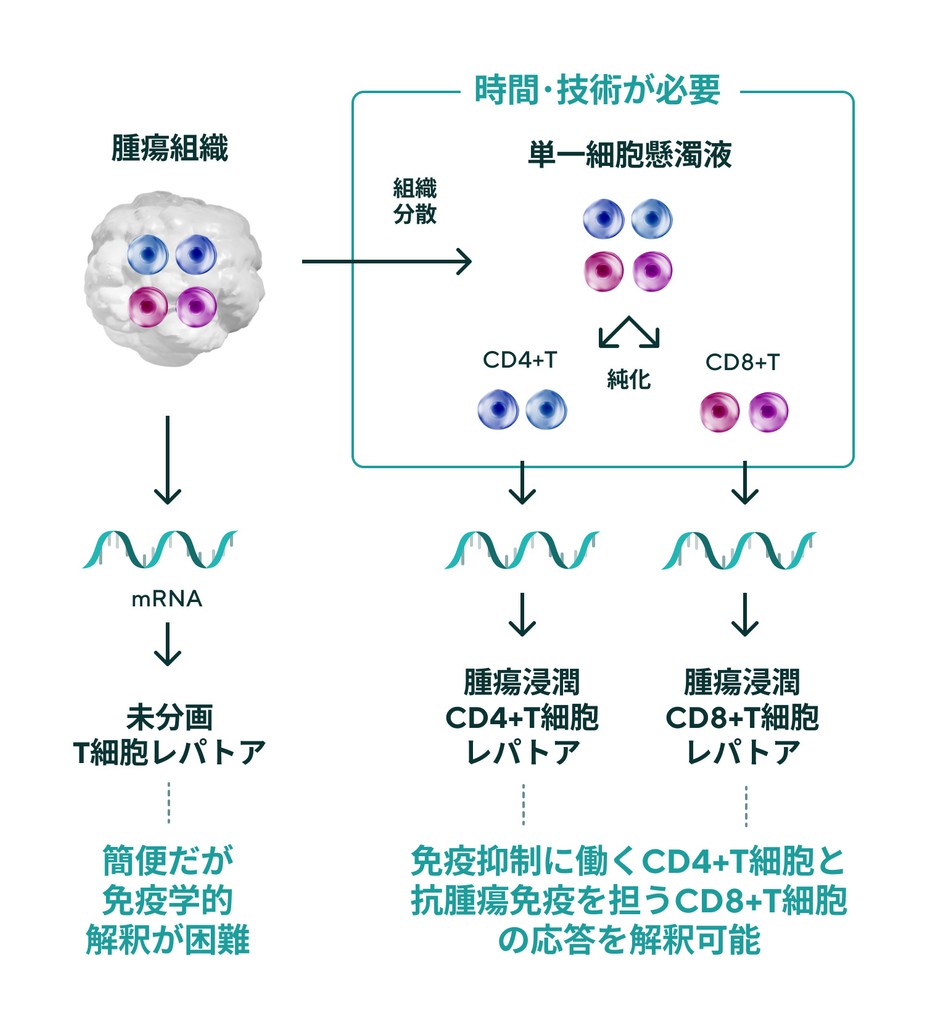
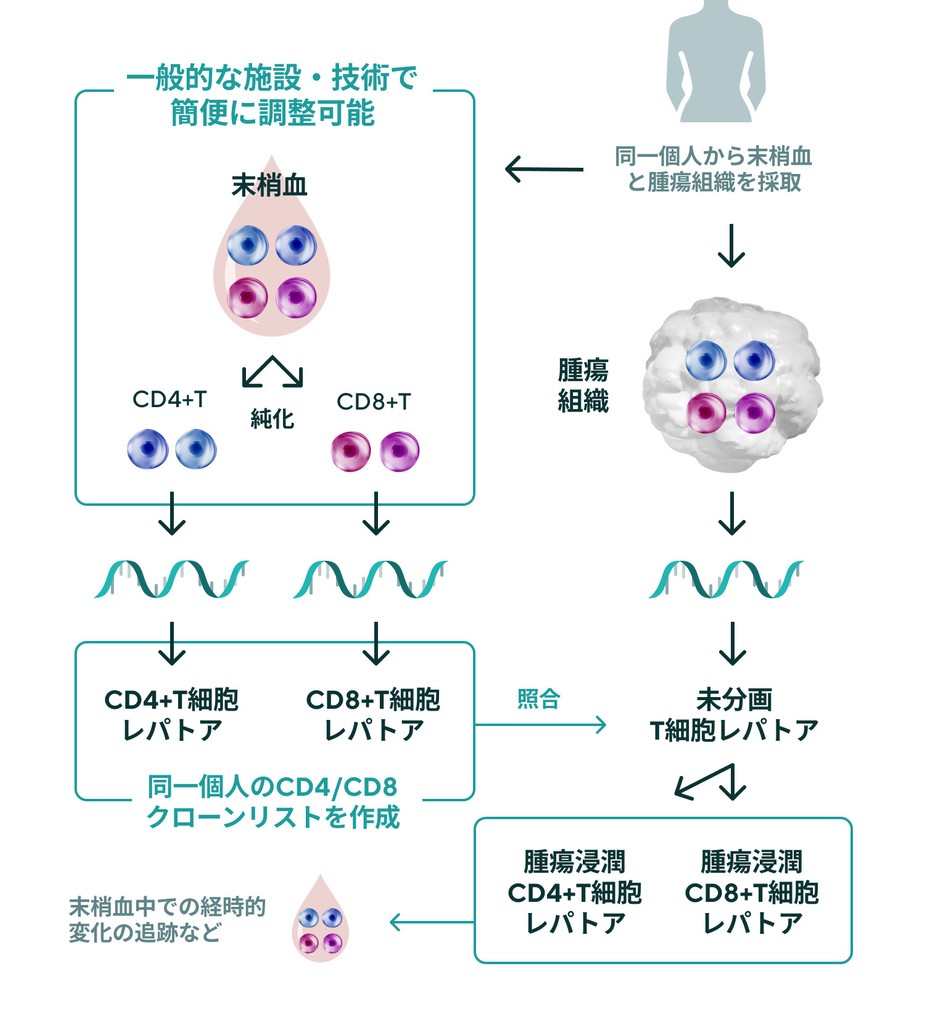
Fractionation of CD4+ T cells and CD8+ T cells is critical for TCR repertoire analysis. On the other hand, purification of T-cell subsets from cancer biopsy tissue and other sources is a significant technical and time burden. IGT has developed a duplicate clone analysis technology (patent pending) that generates a list of clones belonging to CD4 or CD8 based on the TCR repertoire of peripheral blood CD4+ T cells and CD8+ T cells of the patient and determines whether the clone present in the TCR repertoire of unfractionated tumor tissue is CD4 or CD8.
This duplicate clone analysis technology allows for easy analysis of changes in tumor-reactive CD4+ and CD8+ T-cell clones before and after treatment or over time.
* Fractionation options for CD4 and CD8 T cells from peripheral blood mononuclear cells are also available.
Peripheral blood TCR analysis by other companies' conventional methods
Tumor-associated T cells are only present in a small portion of the peripheral blood, making difficult to assess.
TCR analysis fpr peripheral blood and tumor duplication by IGT
Any facility can send peripheral blood and tumor biopsies collected at diagnosis to sent to IGT to evaluate specific anti-tumor T-cell responses.
Conventional tumor-infiltrating T-cell leptorenalysis
Simple but not immunologically interpretable.
Time and special techniques are needed to enable immunological interpretation.
Tumor and peripheral blood duplicate clone analysis technology for
versatile tumor-infiltrating CD4/CD8T cells repertoire analysis
Information analysis
* Option to have a T cell immunology expert advise you on sample setup and analysis according to your research objectives.

mRNA capture and reverse transcription

Adapter addition
Double-stranded DNA synthesis

TCR Gene Amplification

TCR library for NGS
How to request
Sample acceptance
・
Purified total RNA
・
Cell lysate dissolved in our designated lysis solution
・
・
Unfractionated frozen cell samples (Contracted by igt from T cell preparation onward)
・
Cryopreserved tissue specimens (IGT is entrusted with the preparation of cell suspensions and beyond)
Send samples to
〒277-0882
千葉県柏市柏の葉6丁目6番2号
三井リンクラボ柏の葉1 304号室
TEL:04-7192-8732
ImmunoGeneTeqs, Inc.
How to send samples
・
Please make sure that the sample is dry and leak-free, and send it to the following shipping address (We can accept samples until 5pm on weekdays.)
・
If the sample to be sent is RNA, please send it frozen with enough dry ice to keep it frozen.
Lead Time
From sample receipt
2.5 months
Deliverables
・
Work report
・
Complete sequence data
・
Mapping result files, etc.
Reference price
Number of samples
Price (incl. tax)
1
¥99,000
4
¥360,800
8
¥642,400

If you would like to request an analysis, please fill out and submit the hearing form at
We will prepare a quotation based on the information you send us.
Clonal spreading of tumor-infiltrating T cells underlies the robust antitumor immune responses
Transient Depletion of CD4+ Cells Induces Remodeling of the TCR Repertoire in Gastrointestinal Cancer
First-in-human phase 1 study of IT1208, a defucosylated humanized anti-CD4 depleting antibody, in patients with advanced solid tumors
TCR Repertoire Analysis Reveals Mobilization of Novel CD8+ T Cell Clones Into the Cancer-Immunity Cycle Following Anti-CD4 Antibody Administration