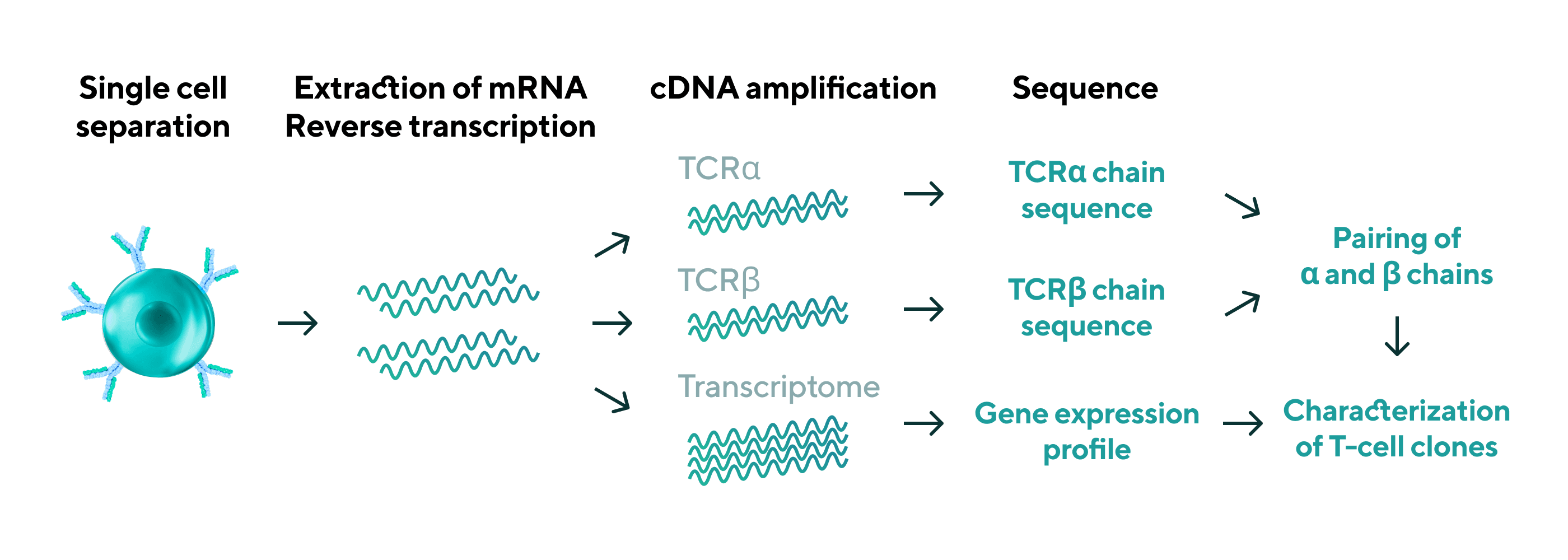
By integrating scRNA-seq and TCR repertoire analysis, gene expression information and TCRα/β pair sequences necessary for clone reconstitution can be simultaneously analyzed for individual T cells. Based on the characteristics and frequency of each clone, it is possible to narrow down clones that are promising for TCR gene therapy and other applications.
* For TCR repertoireanalysis, please also refer to the TCR repertoire analysis page. Here, TCR repertoireanalysis is performed at the single cell level.
・
Single-cell RNA-seq is available with either whole transcriptome amplification (WTA) or targeted sequencing using BD's Immune Response Panel.
*Target sequencing can optionally analyze genes not included in the target panel.
・
Capable of up to 20,000-30,000 cells/analysis, pairing efficiency of mouse and human bio-derived TCRα/β is around 20%-80% (depending on cell activation state).
・
Simultaneous analysis with protein using BD's Abseq, BioLegend's TotalSeq, etc. is also available
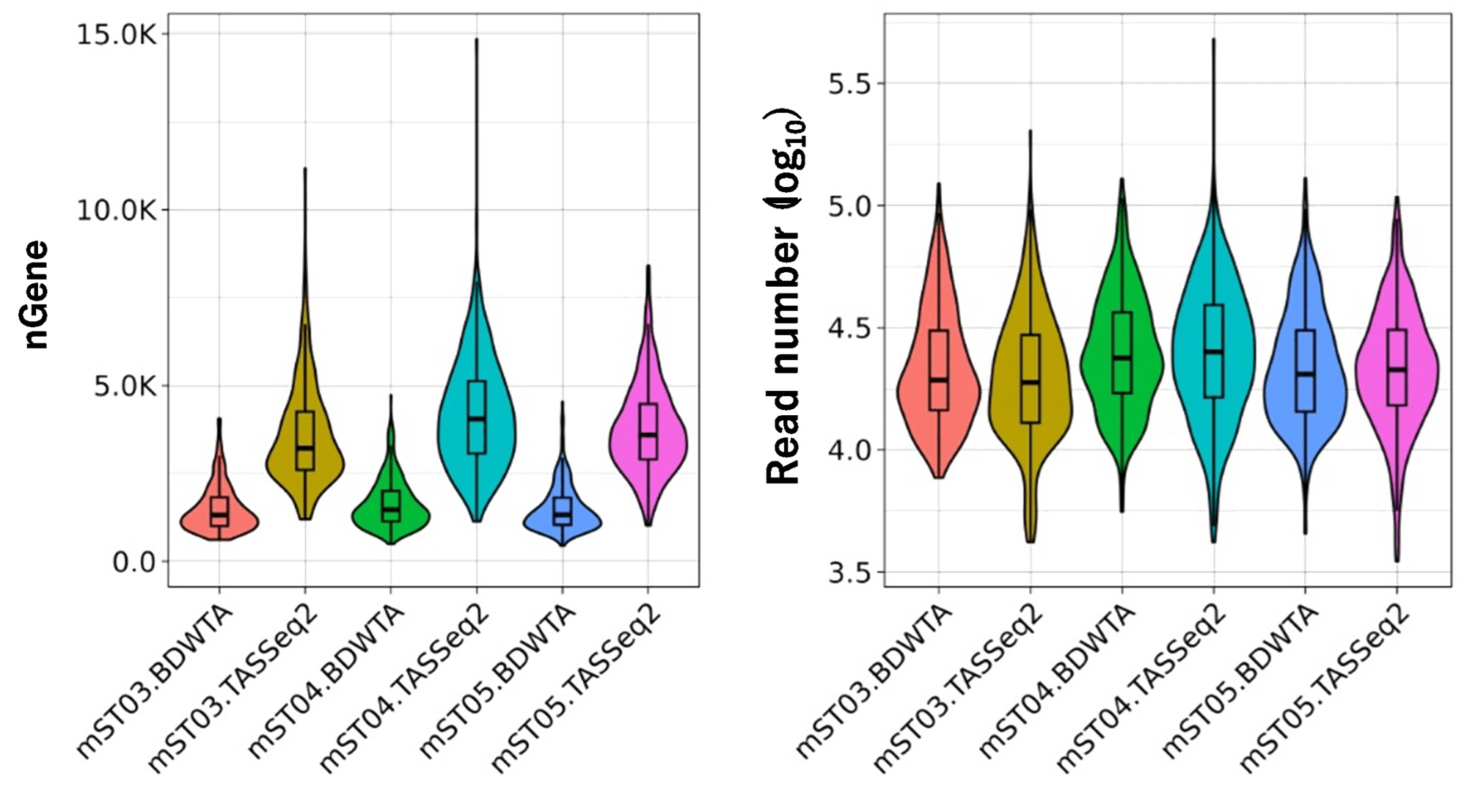
TAS-Seq2 detects about 3 times more genes than BD's WTA kit
TAS-Seq2 has a higher T-cell receptor pairing rate than BD's WTA kit
Example of analysis of tumor-infiltrating CD8⁺T cells in a B16F10 subcutaneously implanted tumor mouse model upon anti-PD-L1 treatment (WTA)
Approx. 50,000 reads/cell
EXPRESSION PATTERNS OF REPRESENTATIVE CD8+ T CELL MARKER GENES
BD WTA
Clusters identified by TAS-Seq2
0: terminally exhausted, 1: effector, 2: proliferated, 3: exhausted progenitor-1, 4: exhausted progenitor-2, 5: naïve-like, 6: stem-like, 7: dying cell
・
TAS-Seq2 can identify CD8+ T cell subsets that are difficult to identify with BD's WTA kit
・
TAS-Seq2 can clearly detect genes involved in the regulation of CD8+ T cell function with a lower drop-out rate
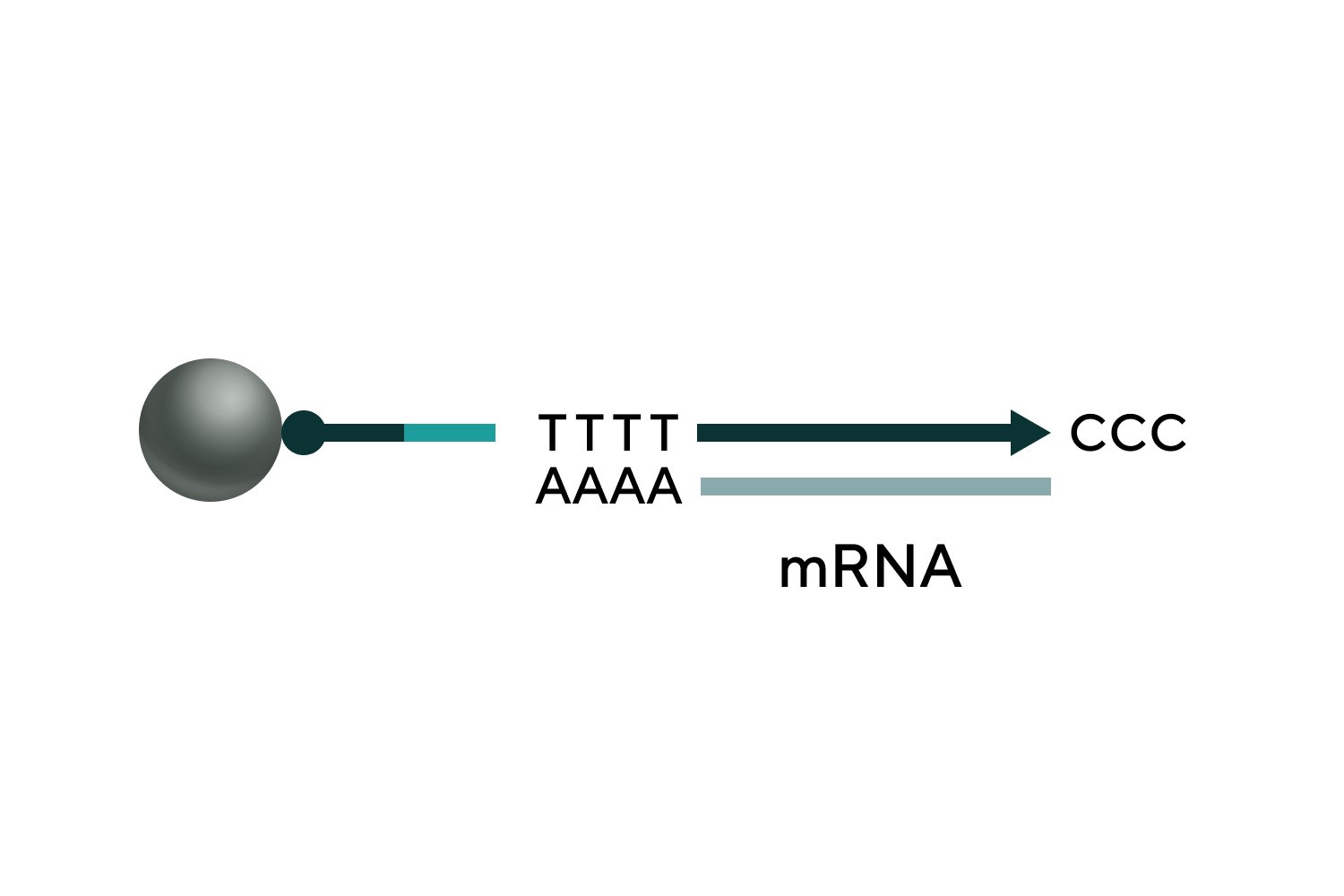
Reverse transcription
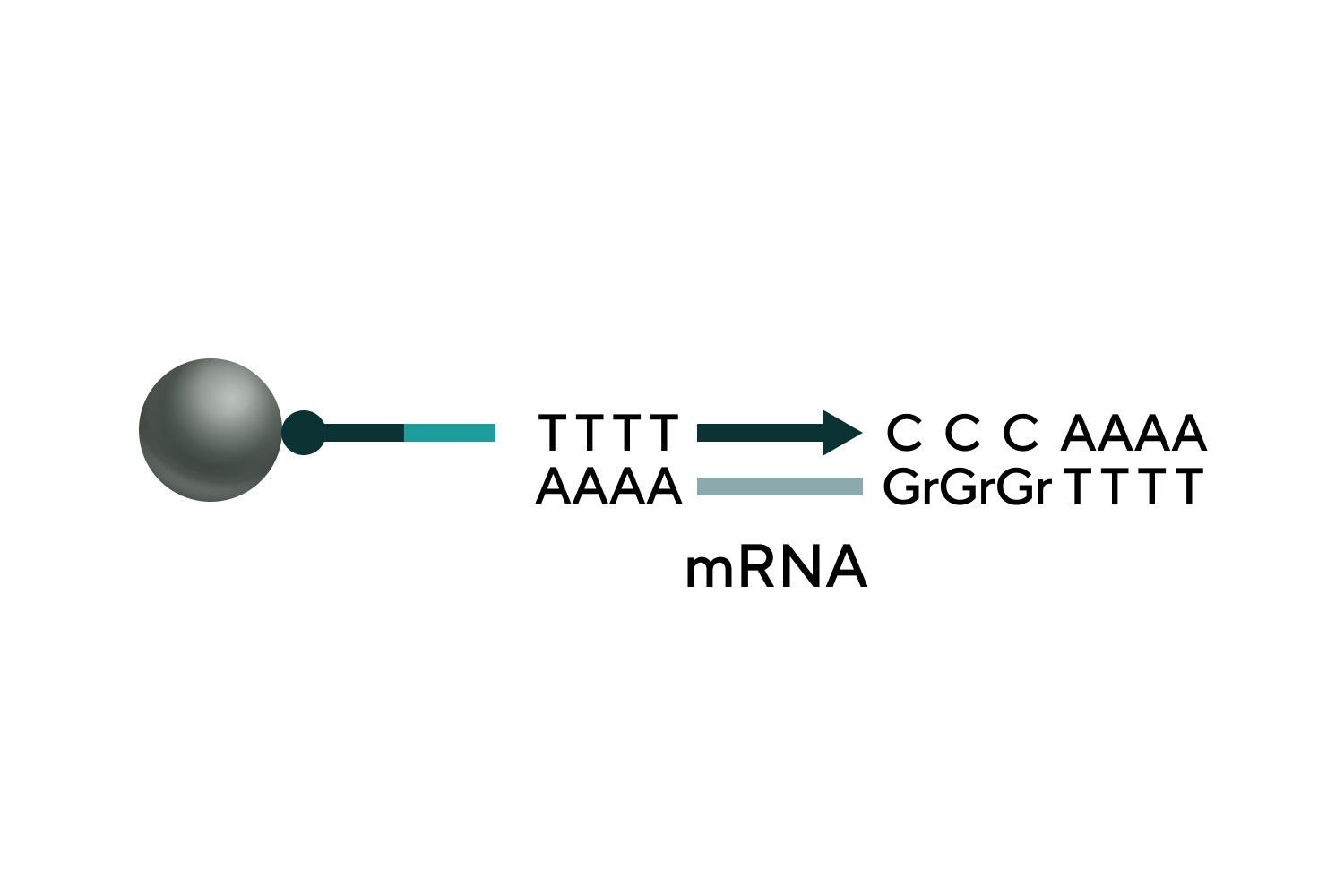
PolyA addition to the 5' end
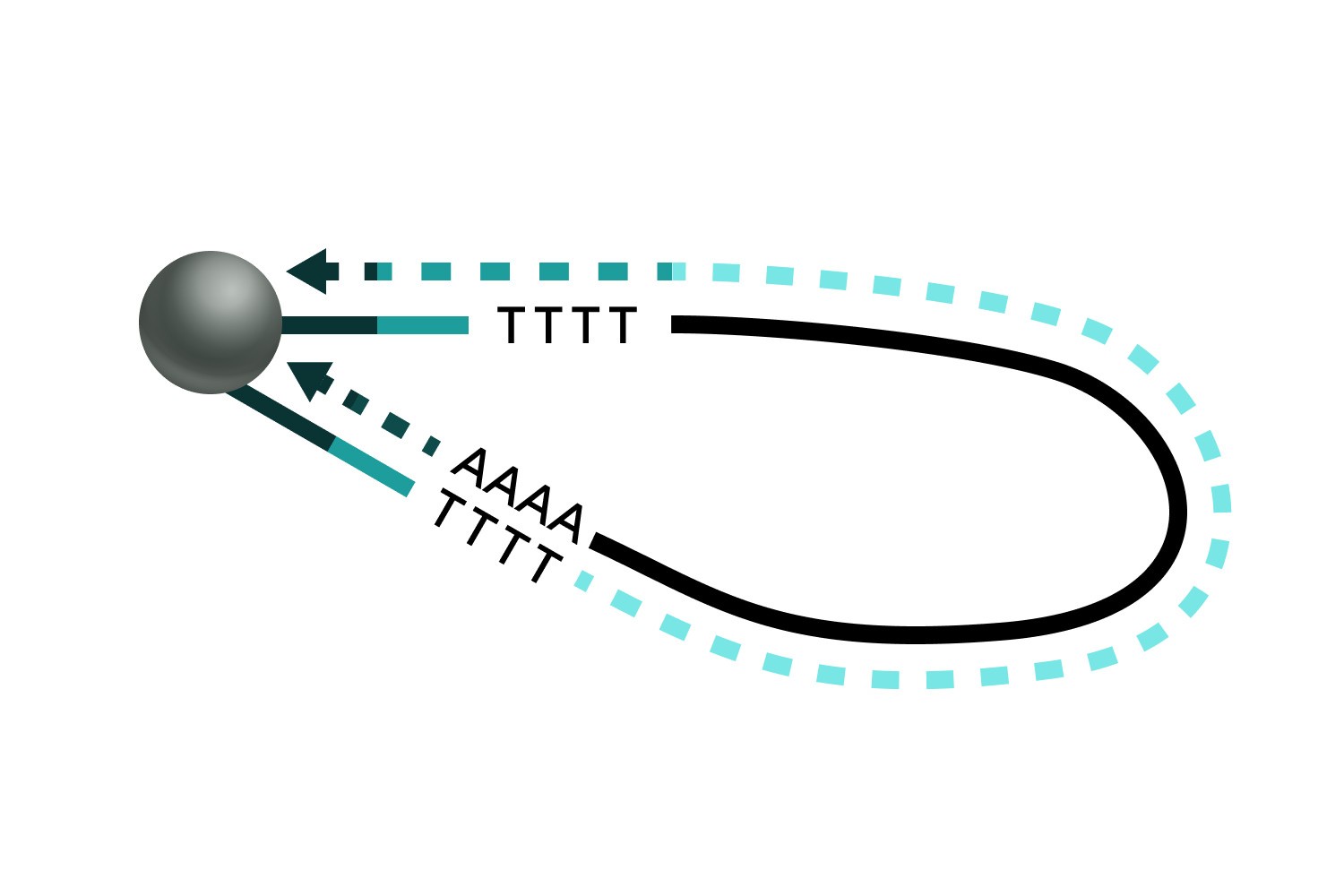
Self-hybridization and elongation
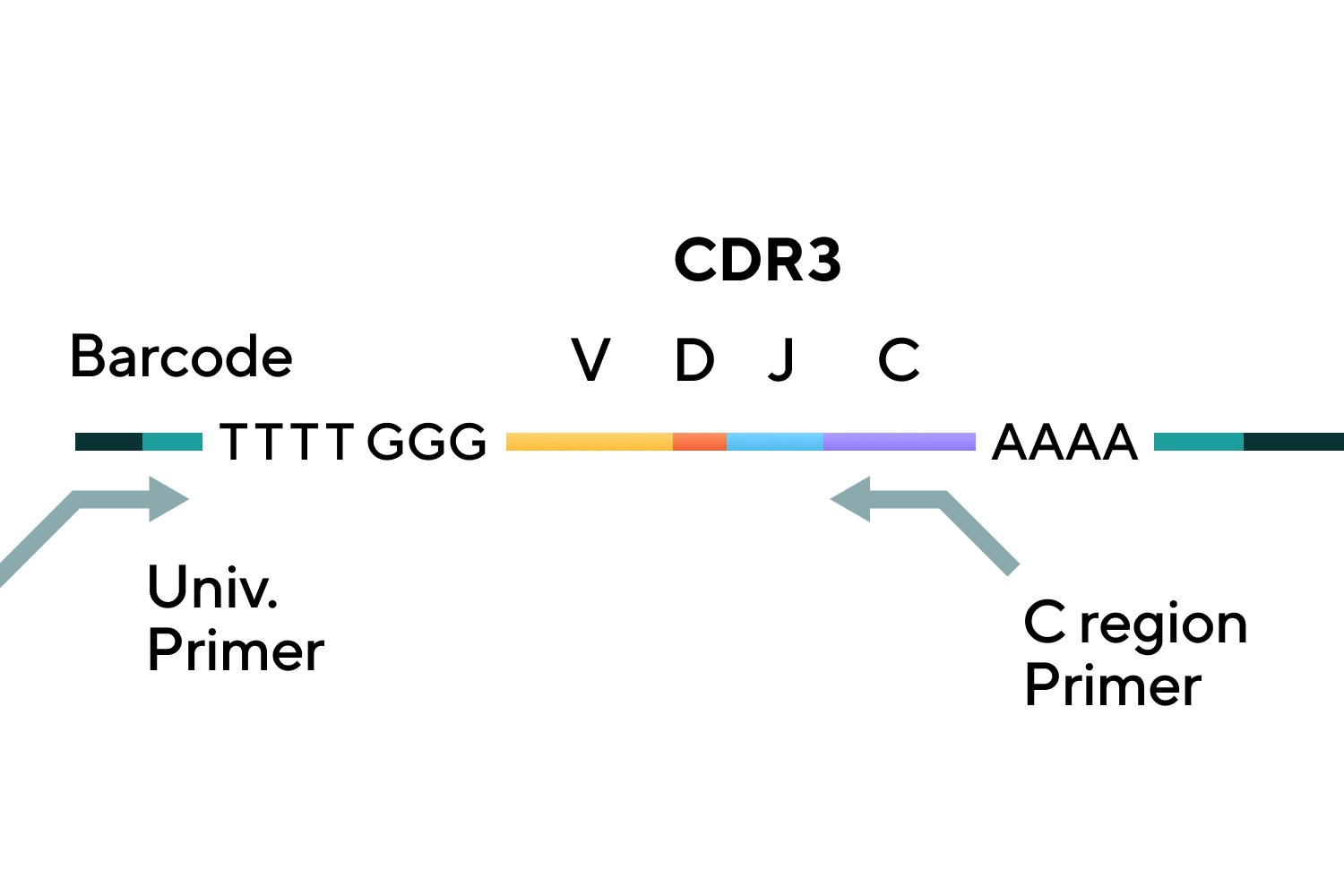
TCR Gene Amplification
TCR a/b library
Matters to be checked in advance
* For multiplex analysis, please refer to the FAQ page "About Multiplex Analysis".
How to request
Sample acceptance
・
cDNA synthesized by our specified protocol (IGT will undertake cDNA amplification and subsequent steps)
・
Pre-fractionated frozen T cell specimens (cDNA synthesis and beyond contracted by IGT)
・
Unfractionated frozen cell specimens (IGT is entrusted with t cell preparation and beyond)
・
Cryopreserved tissue specimens (IGT is entrusted with the preparation of cell suspensions and beyond)
* When cryopreserving cells and tissues, please use the preservation solution and freezing method specified by us.
How to send samples
・
Please make sure that the sample is dry and leak-free, and send it to the shipping address below. (Available until 5:00 p.m. on weekdays)
・
If shipping frozen tissue or cells, please include enough dry ice to maintain the frozen state and ship frozen.
・
Please refer to "Sample Shipping Address" for shipping address.
* *Except for remote areas, please send by next morning (except Saturdays, Sundays, and national holidays).
Send samples to
〒277-0882
千葉県柏市柏の葉6丁目6番2号
三井リンクラボ柏の葉1 304号室
TEL:04-7192-8732
ImmunoGeneTeqs, Inc.
Lead Time
From sample receipt
2.5 months
Deliverables
・
Work report
・
Complete sequence data
・
Mapping result files, etc.
The official report will be delivered on a hard drive at the time of delivery
A mapping analysis report will be included in the formal report
An example of the report can be seen in the example of the analysis of mouse tumor-infiltrating T cells
scTCR-seq (WTA)
Reference price
1
20,000
2
20,000
4
8
5,000
Example of analysis of mouse tumor-infiltrating T cells
Tumor-infiltrating T cells (14,100 cells) were classified into 10 clusters and visualized by dimensional compression; clones sharing the TCR were then defined and the properties of each clone were evaluated; data consisted of single cell RNA-seq data and single cell TCR repertoireanalysis data.
The actual report will also be in this style
Authors:Hiroyasu Aoki, Masahiro Kitabatake, Haruka Abe, Peng Xu, Mikiya Tsunoda, Shigeyuki Shichino, Atsushi Hara, Noriko Ouji-Sageshima, Chihiro Motozono, Toshihiro Ito, Kouji Matsushima, and Satoshi Ueha
Journal, 掲載日:Cell Reports, 2024年3月8日

If you would like to request an analysis, please fill out and submit the hearing form at
We will prepare a quotation based on the information you send us.